ORTRD Tutorial: How to Explore Gene-Transcription Factor Regulatory Networks
1.1 Search
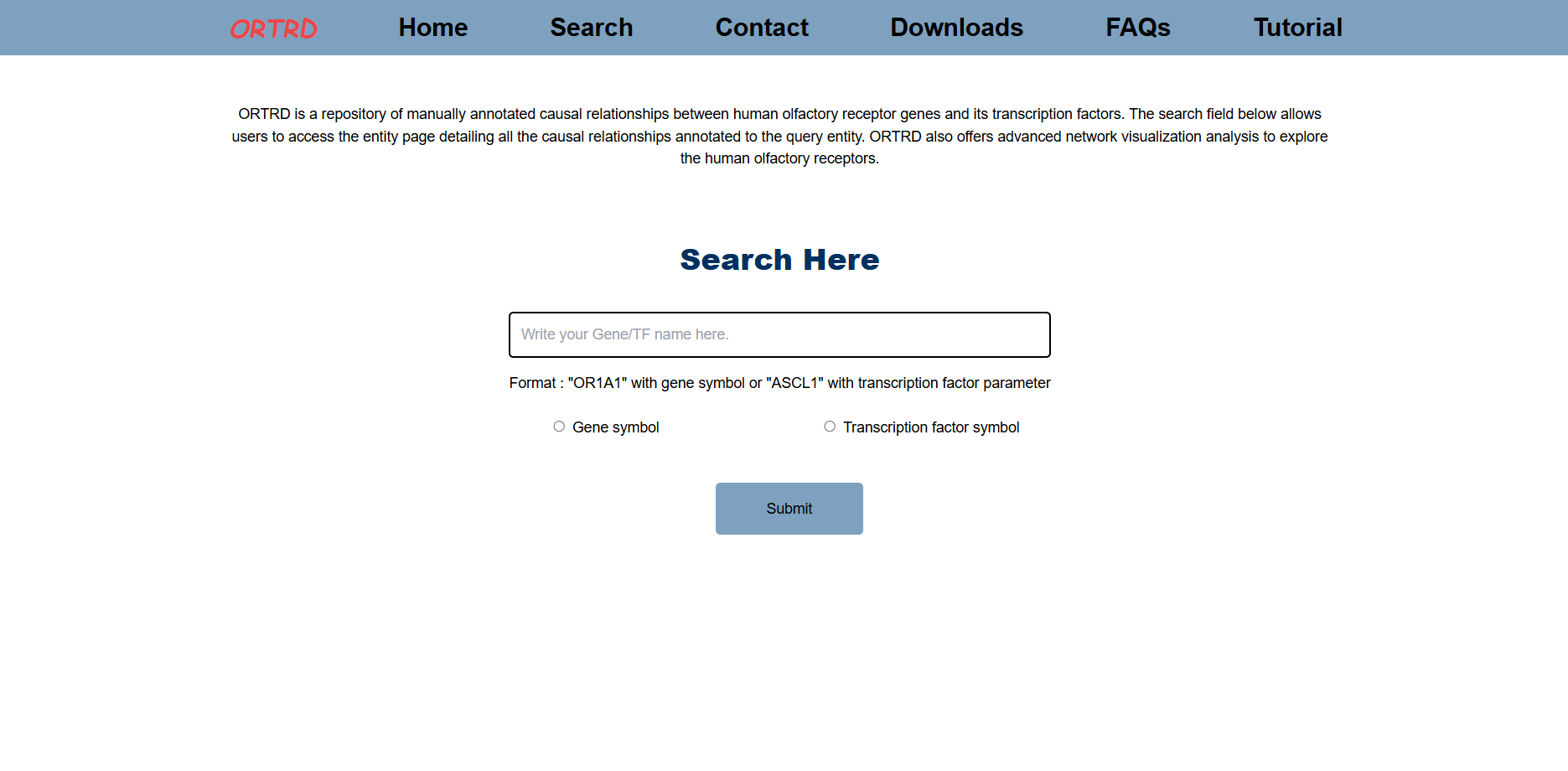
To begin, open the Home page and click on the Search option in the header. ORTRD supports two types of searches:
- Search by Gene Symbol
- Search by Transcription Factor Symbol
Enter a single search term in the search box, choose the appropriate category, and click on Submit. Results will be shown as a table containing matched entries on the browse page.
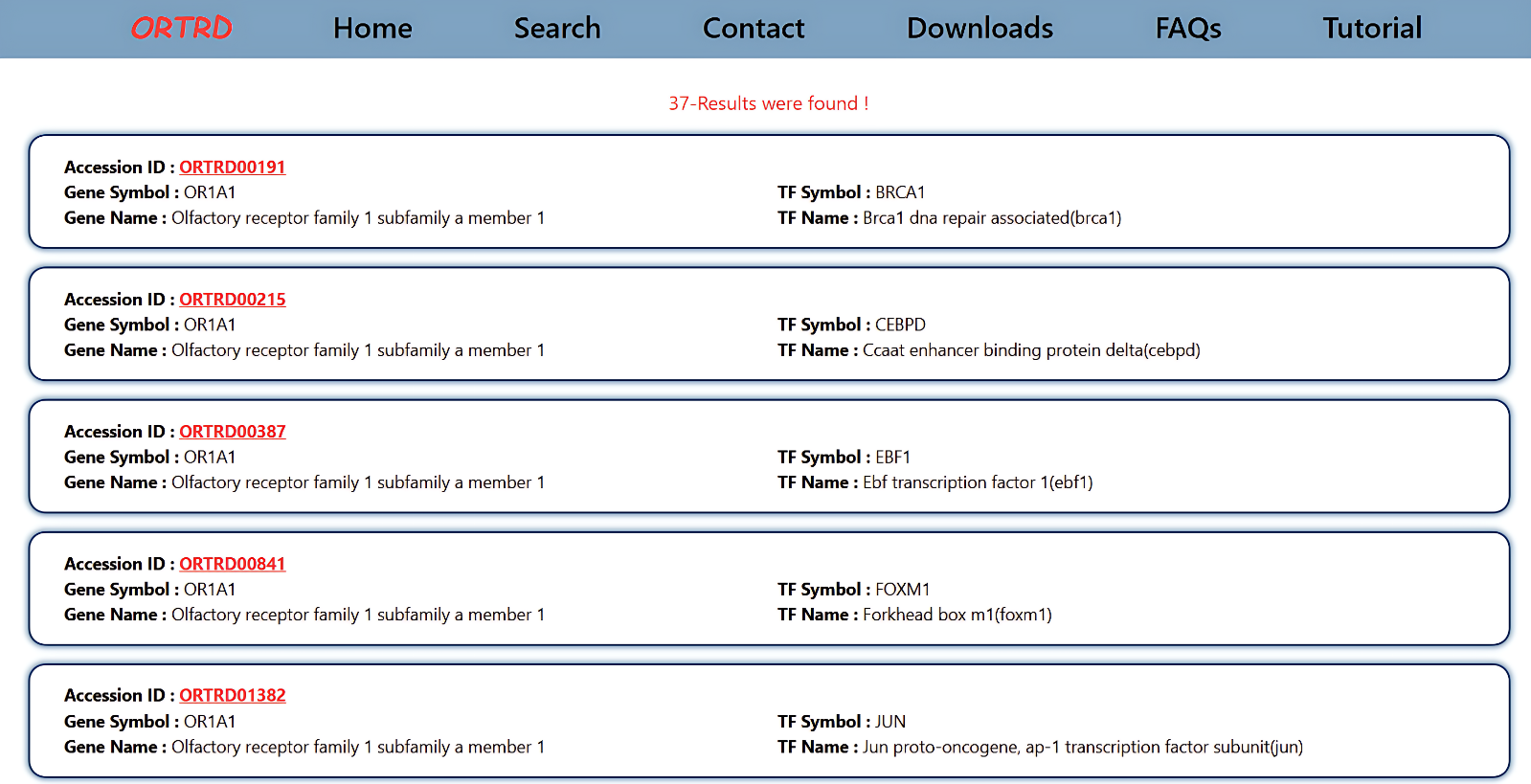
1.2 Browse
The Browse page displays the data in an interactive card layout. Each card includes:
- Accession ID
- Gene Symbol
- Gene Name
- Transcription Factor Symbol
- Transcription Factor Name
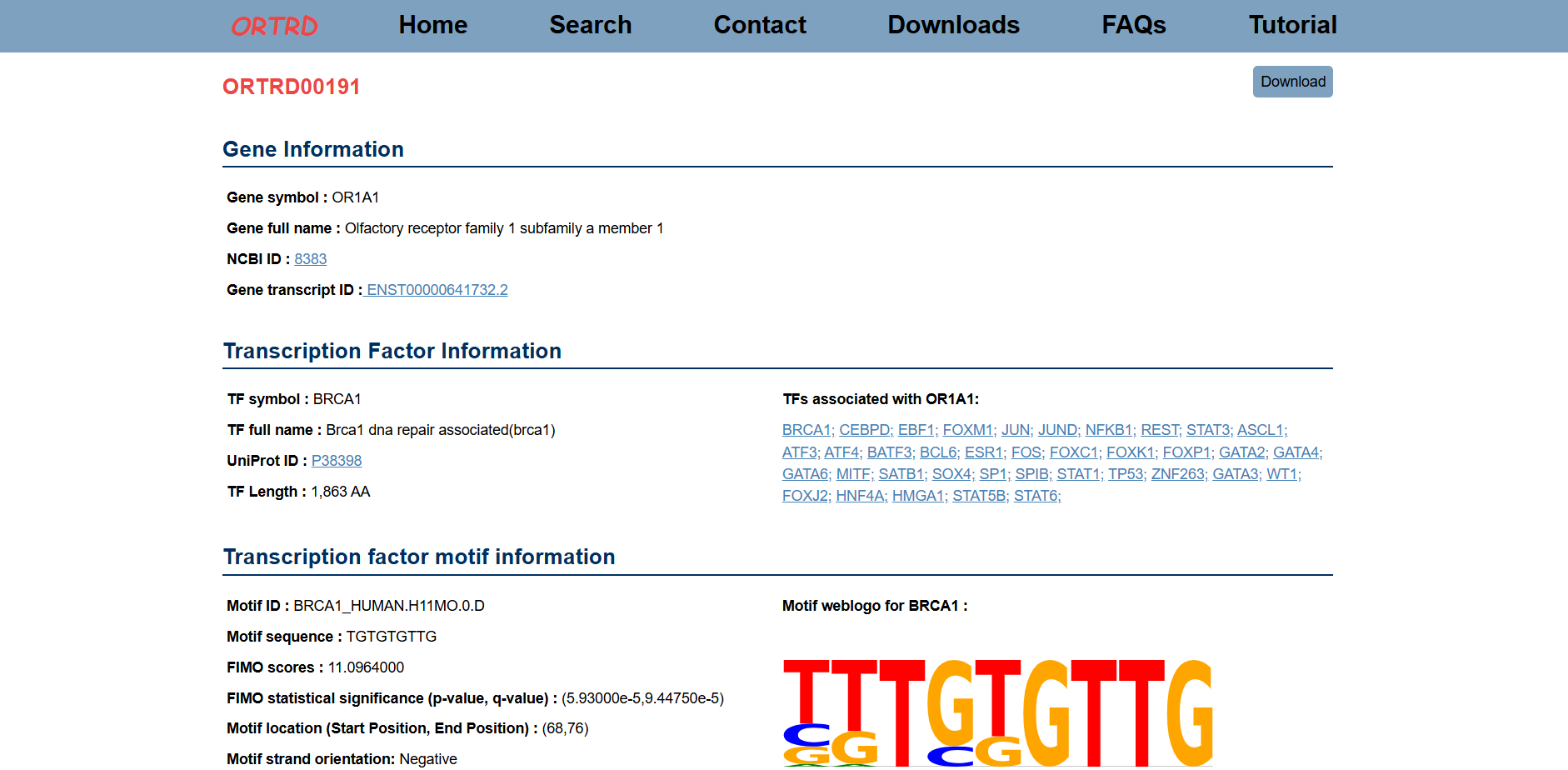
Users can click the Accession ID (e.g., ORTRD00191) to view detailed information about the selected dataset.
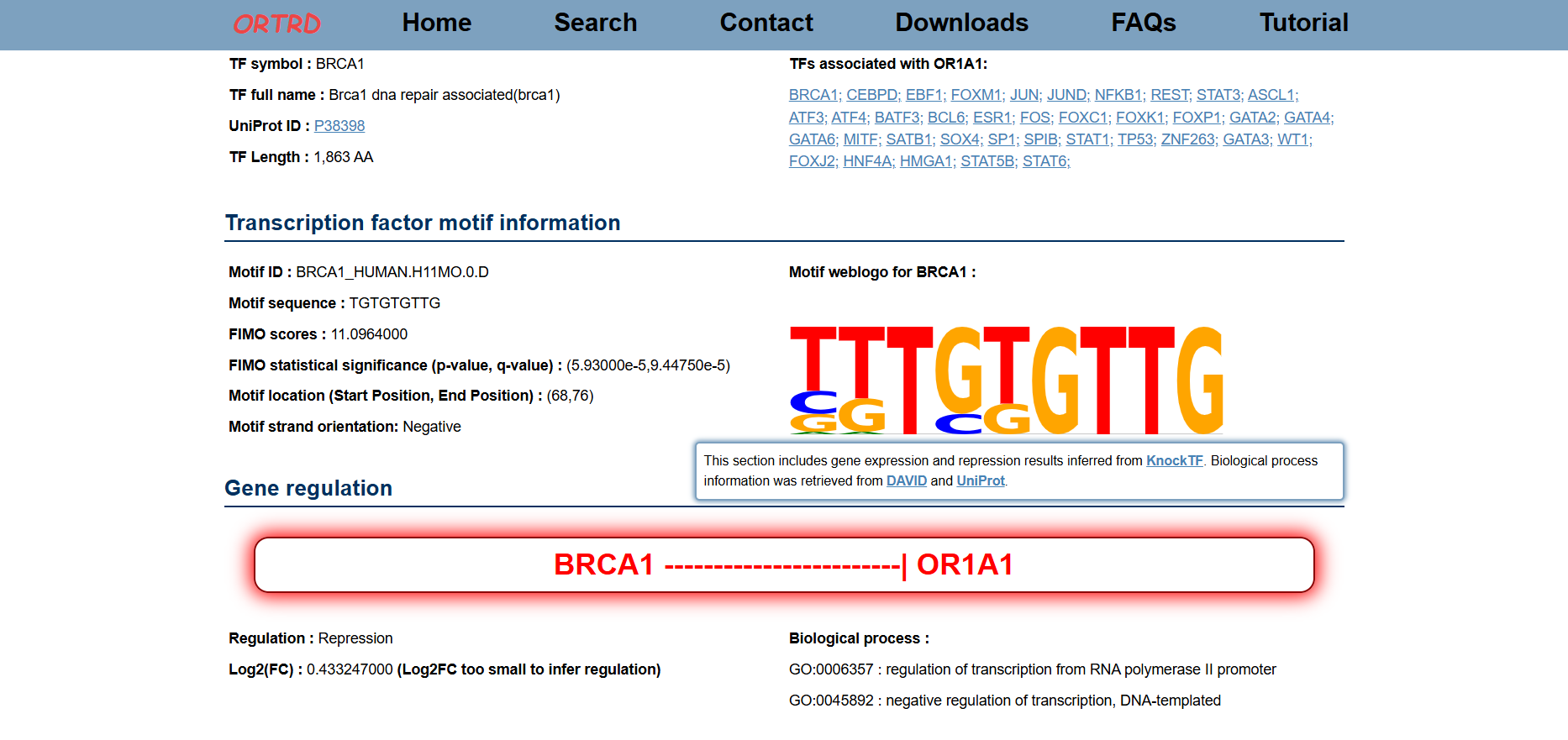
1.3 Result
After clicking on an accession (e.g., ORTRD00191), the Result page will display:
- Gene Information (e.g., OR1A1)
- Transcription Factor Information (e.g., BRCA1)
- Motif Information
- Gene Regulation (e.g., Repression)
- Associated Gene Regulatory Network (GRN)
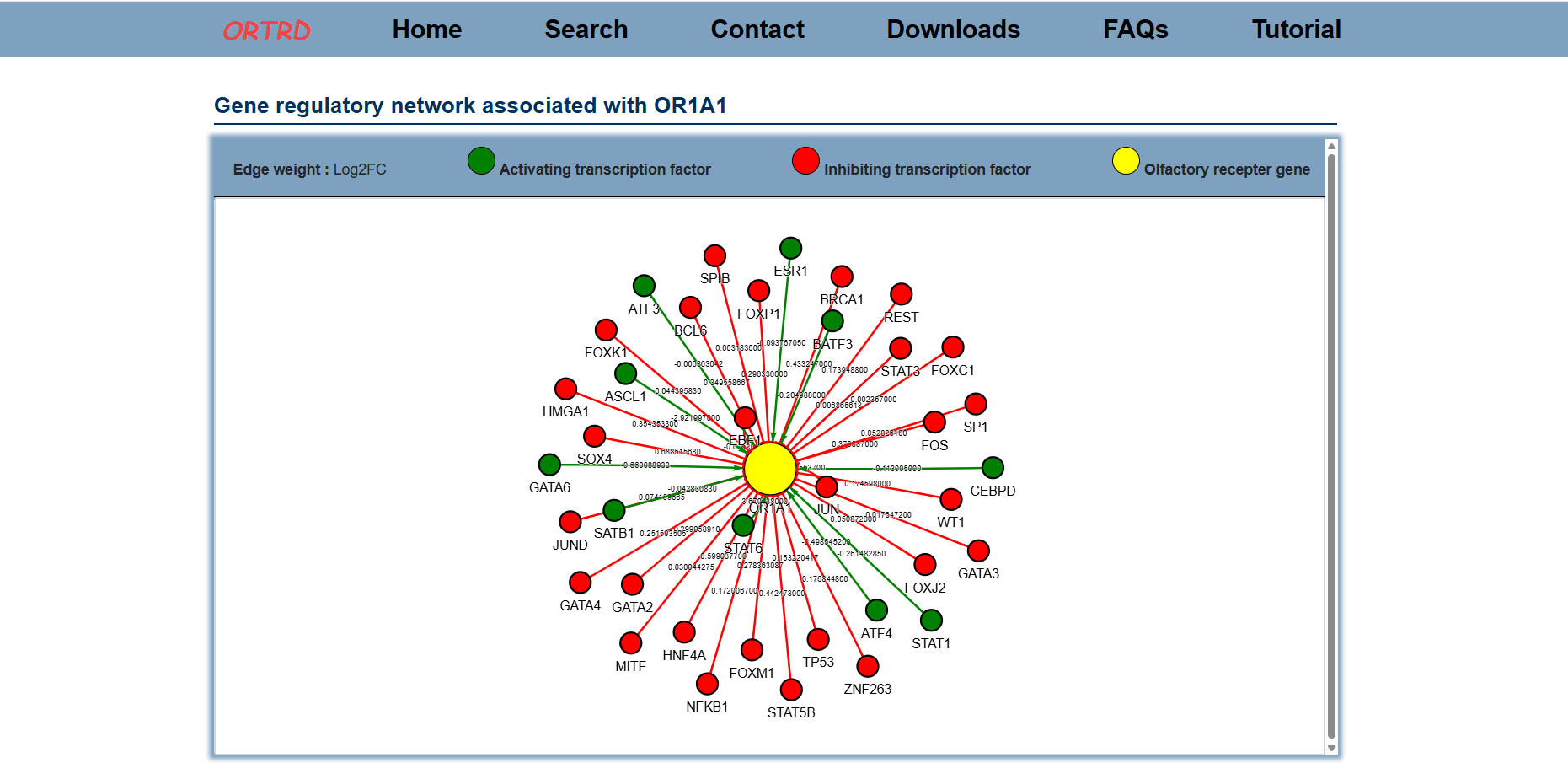
1.4 Network Analysis
ORTRD constructs Gene Regulatory Networks (GRNs) by integrating olfactory receptor genes with their transcription factor interaction data.
Users can input either a gene symbol or a transcription factor symbol to visualize the Gene-Transcription Factor Regulatory Network. Target gene-TF pairs supported by ChIP-Seq data are highlighted with bold edges, and network strength is represented by Log2(FC) values.